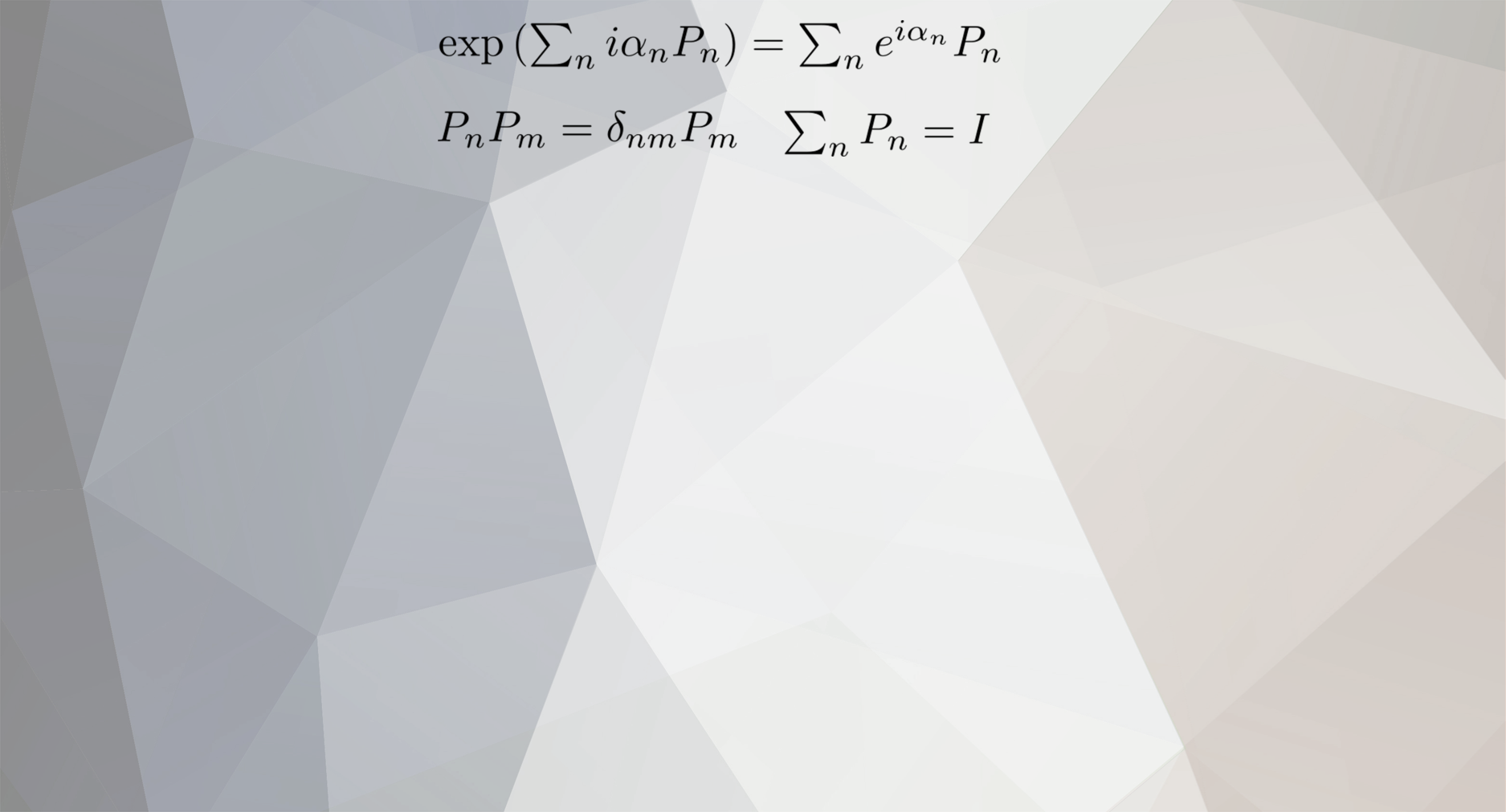-
Posts
4785 -
Joined
-
Days Won
55
Content Type
Profiles
Forums
Events
Everything posted by joigus
-
I've read the premises of your paper. I don't understand any of it. In particular, assuming a closed differential manifold is going to give you a lot of problems with boundary conditions. Maybe that was on purpose. I do agree on one fundamental point, though. It is where you say, "It is cheaper to not get excited than to get excited." I suggest you follow your own intuition there.
-
The way I see it, the problem with 'God caused the big bang' is that there's a theory that, as it stands, is trying to grope further back from the big bang with a certain amount of success. If it does succeed, God will have to step further back. If God doesn't want it to succeed, who's going to tell him, 'please, can you step back a little bit?, we can't quite see out there.' It's a god that seems to install himself in the hollows of knowledge. As to intelligent design, I see it as just a fancy name for wishful thinking. Who designed oncogenes? Errors in meiosis that lead to birth defects? Have you seen a person with microcephaly? If anything, biology suggests to me that there is no design nor there is a designer. No engineer would design a machine that uses the same material for fuel, structure, communication network, and then throws some of it away while it's recycling part of it, and needing more! This 'designer' would also have to intentionally make his machines go to waste just as they come out of the production line every so often, just for the fun of it. There goes my machine, oops!! So it's also a god who doesn't seem to care. Having said that, I do find much of value in Taoism, which most people think is a religion, but I don't see it that way. It is a practical philosophy, it predicates the 'I don't know,' which is very healthy. The practice of Zen, e.g., is a breathing technique and a constant question 'what is "I".' And it doesn't require God. Only problem (for me) is I'm not very keen on tunics or prayers or looking at the statue of anybody as something particularly inspiring. (Although I love the ringing bells.) Maybe that's why I say 'Taoism' and not 'Buddhism.' Bias? Perhaps. But we're all biased.
-
I never said that. The bouncing photon clock that you talked about involves photons interacting with matter, which no longer is a photon travelling through the gravitational field. Such system, with two parallel mirrors and a photon bouncing back and forth, doesn't work like you claim it does. If you ever see a radiation pressure fan or radiometer, you will understand what I mean just looking at it. You won't have to think or listen to anybody, or read what they say, which for you is a definite advantage: photons push against a mirror, you see? That's why your idea didn't work. But I'm starting to lose track of what you're saying. You've talked so much nonsense today I can't keep track. Now those are real photons, not the ones that are in your mind. Cheers
-
Yes, this is Richard Feynman in 1965. Have you heard of entropic gravity? Erik Verlinde deduces Einstein's equations and Newton's laws. And it is by no means sure it is the right theory: https://en.wikipedia.org/wiki/Entropic_gravity#Criticism_and_experimental_tests IOW, people don't buy it just yet. Why should scientists pay more attention to you than to Verlinde, for example? Com'on, don't make a fool of yourself any longer. This is really painful to witness.
- 344 replies
-
-1
-
rjbeery, I'm giving you some homework: https://www.amazon.com/Introducing-Einsteins-Relativity-Ray-dInverno/dp/0198596863 You don't sound to me like you're completely off your rocker. Maybe you've tried to get into the forest a little too deep, a little too devil-may-care, and without dropping your breadcrumbs on the floor. I don't want to be completely negative. So there you are.
-
When a particle is massive, wavelength has to do with mass by De Broglie's relation, \[p=mv=\frac{h}{\lambda}\] p is called 'momentum.' m is the particle's mass, v is the particle's velocity, \lambda is the wavelength and h is Planck's constant. For photons though, it's also, \[p=\frac{h}{\lambda}\] But expressing p as mv is no longer valid. So the photon's wavelength has nothing to do with its mass.
-
1) I do not believe anything, I need, demand AAMOF in this context, logical proof or experimental evidence. You have neither. 2) Clock rate is a relative (frame-dependent) quantity. Plus you only too obviously don't understand special relativity, let alone GR.
-
Before re-iterating too much, I would suggest you re-read what people are telling you, and then re-think for a change.
-
OK, but you're drifting away from entropy. I mean, electrons, photons or pi mesons can 'experience' curvature (and there I do accept your term,) but not entropy, as entropy is a property of your level of description. It's to do with lost information, and particles don't lose any information AFAIK. Or the concept of them 'experiencing loss of information' doesn't seem a reasonable physical concept. What entropy growth (or information loss) has to do with is a quite abstract but useful concept that is called 'volume of phase space.' It is a measure of the amount of information that a physical system contains just because of the fact of being in a certain dynamical state. This 'volume of phase space' is neither lost nor gained; it's constant. Just constant. Entropy is the part that is hidden to my description. We could say, \[S=\textrm{constant}\] This is sometimes called 'microscopic entropy' and its conservation is the most fundamental physical principle there is. Now, it just so happens that many things go on without us knowing about it. Only because there is a fiduciary value of a quantity that stays constant and I can associate with the information content of a system, can I speak about loss of information. Otherwise it wouldn't make any sense. I hope that helps to clarify the situation as to the entropy. It's a really confusing concept, and the great mathematician John V Neumann once said that physicists don't know what entropy really is. No longer the situation, I think.
-
I'm not sure whether this is related to the problem you're having, but your oxidation looks to me like an reduction. The magnesium is capturing electrons.
-
How do you define 'absolute'? For any object moving, you set clocks and systems of laser beams going back and forth to measure positions and time intervals (the latter calculated taking into account how much the signals delay in reaching the observer, it's not 'subjective' time we're talking about; it's not 'when I see the object.') Now, let's call them dt, dx, dy, dz. The so-called proper time of the moving object, in Special Relativity, and with the proper generalization, in General Relativity too, is, \[d\tau^{2}=dt^{2}-dx^{2}-dy^{2}-dz^{2}\] For what observer? For all inertial observers! Now, that's what I would call invariant (I'd never say 'absolute'.) And in order to do that, you need a system of signals, as they're trying to tell you. You need a way to bring it all together, so to speak. For a photon, the proper time is always zero, so photons have no internal clocks. While, seen from the 'outside' in empty space, they always go c.
-
Sorry, I didn't see the geodesic equation, I didn't see Einstein's equations derived, I didn't see the equivalence principle, I didn't see the Newtonian limit, I didn't see gravitational horizons, FRW or DeSitter universes or any other cosmologies, I didn't see vacuum energy, I didn't see dark matter, or red-shift, I didn't see a thing that even remotely reminded me of gravity, except in the title and Eddington's paper. I suggest you change your mindframe: Try to prove yourself wrong. If you always try to prove yourself right, you're always going to find a way to be right.
-
I'm just trying to engage Studiot. I tried to +1 you at your reminding me of the Klein paradox, but I messed up. Well deserved anyway.
-
The issue of what mass is and what it's to do with space is very deep indeed, and I think it can be discussed at several levels, has acquired successive layers of sophistication throughout history, and I don't think I will be able to answer to the full satisfaction of anybody. I've tried to compile a brief excursion through it that I will separate, so that nobody needs to read it. But it could serve as reference. </start of digression> Brief history of mass There are several concepts of mass, which should really be carefully distinguished. -Inertia: The opposition to acceleration as defined by Newton in the Principia. But this definition is tautological (force, inertia, what comes first?), and Newton could get away with it only because he was incredibly clever and instinctively new how to get out of the tautology by adding auxiliary hypothesis in the force law. -Gravitational mass (Newton again!): The source of the gravitational field. Some people even distinguish between active gravitational mass (source) and passive (test particle.) But we can immediately identify both if we want momentum to be conserved, as otherwise F_12 =/= F_21. -Then comes Newton, with hypothesis that amounts to equivalent pple. (EP) ==> inertia = gravit. charge (define all as 'mass'.) -Ernst Mach: Mass must be a property of the filling-in of the surrounding space in relation to the particle. ==> mass appears to be a 'non-local' property in the sense that it codifies some properties of the whole universe as related to the particle. Any possible local anisotropy for inertia has always tested negative. -Special Relativity: Einstein finds out that what we call mass is really rest energy reasoning in terms of inertial systems. -GR: Einstein gives up Mach's principle as a constructive principle for a theory of gravity and opts for EP, although he remains impressed by its logical clarity to the end of his life. Then comes QFT, which sees mass as a scale-dependent parameter. Safely defined as a constant for the infrared as the concept we all know and love, but blowing up in our faces when we try to go to higher and higher energies (UV limit.) Then comes a symmetry-related problem with the electroweak force (EW) in the Standard Model (SM). In order to model short range interactions people need to give mass to the gauge bosons, because gauge bosons must be massless if gauge symmetry is to be preserved. Higgs et al. realize that a quantum field with 2 complex (4 real degrees of freedom) does the trick. Three of them are swallowed up by the bosons, giving them mass, and the other one must have an ephemeral life as the Higgs. But the thing becomes much more far-reaching when people realize in the laboratory that parity in its different versions (P, CP) is violated maximally all over the place. Dirac's equation connects the left-handed e- with the right-handed e- precisely through the mass term. But what's the right-handed electron doing there? All fermions need some kind of weird scalar companion to give them mass and account for parity violation. So every fermion must be coupled to a Higgs 4-plet. </end of digression> As you see, it's all a mess that English physicists have got us into!! Not in any ordinary sense, no. I agree. Not like a gas, nor like a piece of crystal. I like the way you put it. Yes, that tends to confound things a lot. I actually was quite satisfied when I read the first two answers. Yes! Good point. The Klein paradox: When you consider a particle confined to a place of the size of (half) its Compton wavelength (=twice its mass,) it not longer makes sense to even consider it as a particle, as you are in the particle-antiparticle pair creation regime. I almost forgot. But I think that's an extremely important point.
-
Yes, I know what he means. In thermodynamics and statistical mechanics they are very common terms. Statistical mechanics is only rigorously defined when you make all the extensive parameters go to infinity. It's called the thermodynamic limit. Although I must say that talking about that wouldn't have been my choice, because the question of mass is difficult enough as it is. As I've been mentioned, I'm struggling to add something significant to what has been said. But it's not easy...
-
I've read as much as I've been able to. Basically I think I concur with Mistermack. My take is: Different compilations of very old myths (some going as far back as the origins of agricultural societies) by Judahite intelligentsia, a clique that really started to come in from the cold under king Josiah (c. 640–609) of Judah after the Assyrian domination started waning and the rival kingdom of Israel was no more. King Josiah is known to have declared the finding of a book (believed to be Deuteronomy by most scholars) during some refurbishing works of the First Temple of Jerusalem. Writings and re-writings and juxtaposed interpretations and re-interpretations in many layers were added, culminating in 'final' compilation by priest Ezra, after Babilonian exile at the hands of Nebuchadnezzar II in which some myths show signs of having been fused, e.g., the Adam and Eve narrative. Then that intelligentsia revived under the Persian king Cyrus the Great. Then the books were lost after the Greek, and later Roman, domination. Most sources remaining were Greek translations plus scattered documents in old synagogues, in old Hebrew script. Then the Dead Sea Scrolls appeared in the XXth Century, showing stark contrast in interpretation in certain excerpts, e.g., the 'virgin' attribute of Mary, which is very clearly a mistranslation from Hebrew to Greek. "Ha-almah," the original Hebrew, is translated as "young woman" or "maid," while the word for "virgin," "bethulah," appears nowhere in the originals. Someone picked the Greek word "parthenos," leading to centuries of confusion. The story of Noah and the Flood is probably a re-telling of the story of Uta-Napishtim, from The Epic of Gilgamesh. What archaeology really seems to be telling us is that both the Israelites and Judahites were matter-of-factly polytheists. It's even possible that that went on well into the time of Greek domination by Alexander the Great. Bibliography: The Bible Unearthed Israel Finkelstein & Neil A. Silberman
-
I don't know exactly what you mean with 'experience.' Systems of both massive or massless particles contain entropy, a black hole contains entropy of a very different kind, and even for one quantum particle entropy can be defined in terms of its wave function. Entropy, at the most fundamental level, is defined when the distinctions among different dynamical states are lost. Entropy is the opposite of information. The total entropy of the visible universe in cosmology approximately equals the number of photons, about 10^90. A gas of photons contains entropy. So I suppose the answer to both questions is yes. I've just answered the one that Strange passed on, but he was spot on when he said it's about 'systems.'
-
I was about to tell you about your mistake with the ticking clocks, to do with absorption and re-emission, and elaborating on your messing up red-shift with slowing down. But it would be wasted on you, as there's no one reading at the other end. I close with a quasi self quotation: Gravitational fields do not slow down photons, they just-red shift them and make them bend their trajectories. Read some relativity books. And a literal self-quotation:
-

kinetic energy and black hole formation
joigus replied to rjbeery's topic in Astronomy and Cosmology
T's ok. I just tire easy around morphing arguments. Maybe in other life. -
I meant, -Today I've learnt about: geons Indebted to MigL
-
Gravitational fields do not slow down photons, they just-red shift them and make them bend their trajectories. Think again. In no way does that resemble an electron. A bundle of EM field does not have charge, nor does it invert the sign of its probability amplitude under 2pi rotations, which is required. And I've missed the part where you provide a causal mechanism for gravity.
-

kinetic energy and black hole formation
joigus replied to rjbeery's topic in Astronomy and Cosmology
I cannot be completely sure that the answer has been given, as you seem to falter quite a bit in your premises: Naturally. You must really think what you say, say what you think, and may I add, think what you think. And, No: Covariant under general coordinate transformations. You must really read what you read. -

representing the actual state of the particle by adding up functions?
joigus replied to nec209's topic in Quantum Theory
Well... 1) I don't know who sycamore is 2) I haven't the faintest idea what super-computing power has to do with the OP topic or the follow-ups 3) You must be careful with what I said --in case it's anything at all to do with what you're saying--, because probabilities equal the sum of the probabilities is only true for different (orthogonal) eigenstates. You sound a bit hasty and shouldn't run away with any idea, especially from one thread to another. -

representing the actual state of the particle by adding up functions?
joigus replied to nec209's topic in Quantum Theory
I meant, prob(E_1 OR E_2) = |Amp(E_1)|^2 + |Amp(E_2)|^2 + 2Re(Amp*(E_1)Amp(E_2)) 2Re(Amp*(E_1)Amp(E_2)) is called the 'interference term.' without the 'star' for complex conjugate in the second term and taking the real part. -

Are the weirdnesses of QM still regarded as mysteries to be resolved?
joigus replied to Alfred001's topic in Quantum Theory
Don't try to frame Feynman. He was deceptively clear. As to what really matters, I can't wait to hear your arguments. No, really, I can't wait.