Everything posted by Vahid
-
Cts value confuses me
We buy lyophilized 10CFU standards. They are certified by the manufacturer. How should I check the cover heating activation? in addition, I seal the plate carefully and tighten the sides. We always pipette reverse(which means no bubbles.
-
Cts value confuses me
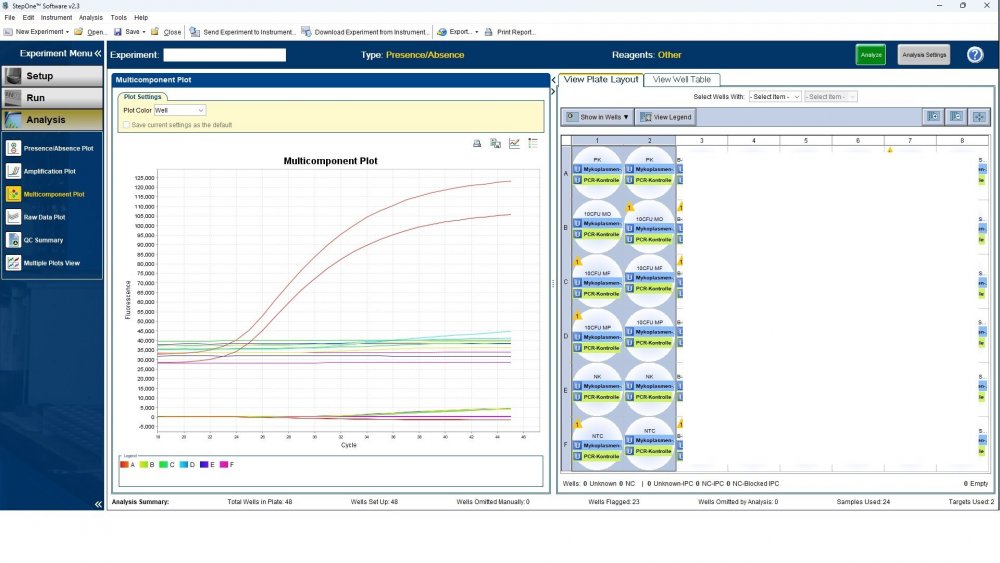
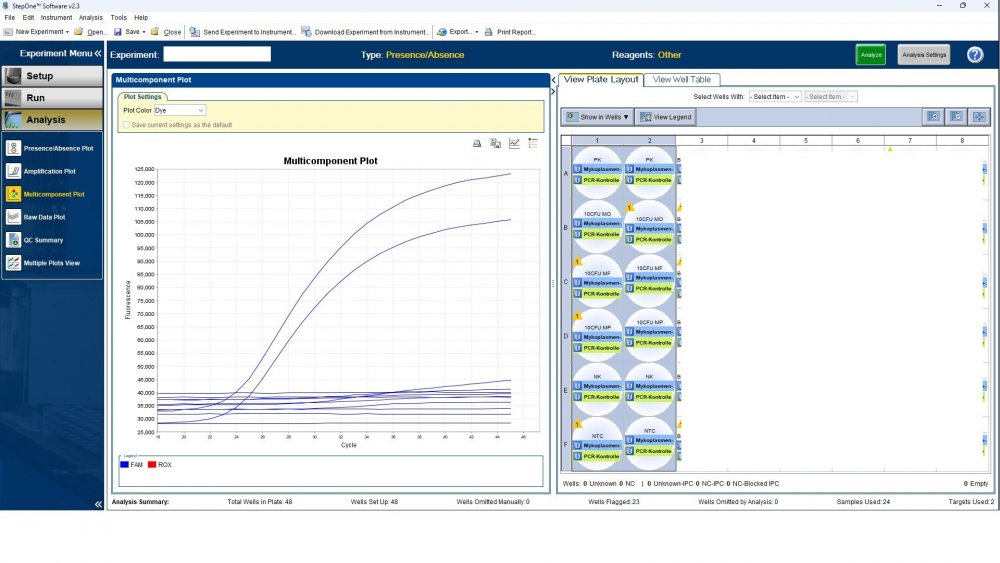
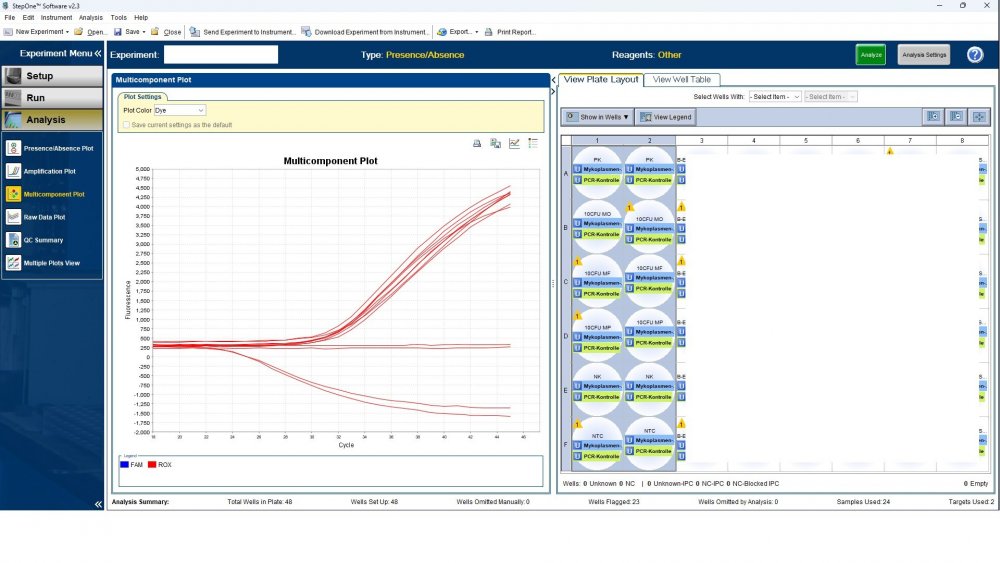
Here is the answers; what you indicate as standard in your list are the extracts from counted cells? Exactly the difference you see is e.g. between B1 and B2, which contains template from the same sample? Yes Is there a trend or is it random? Random As part of your SOP, are you using low-biding filter tips? I uploaded its image. I use Microsart AMP Mycoplasma (Sartorius) to detect the Mycoplasma (FAM, ROX kit) What are you meaning by MM? Attached are pictures of one of the invalid tests.
-
Cts value confuses me
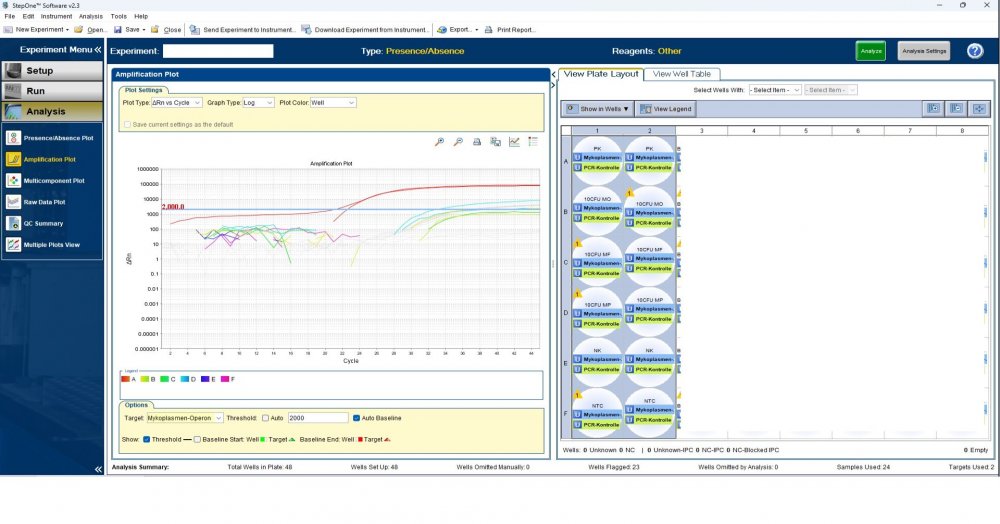
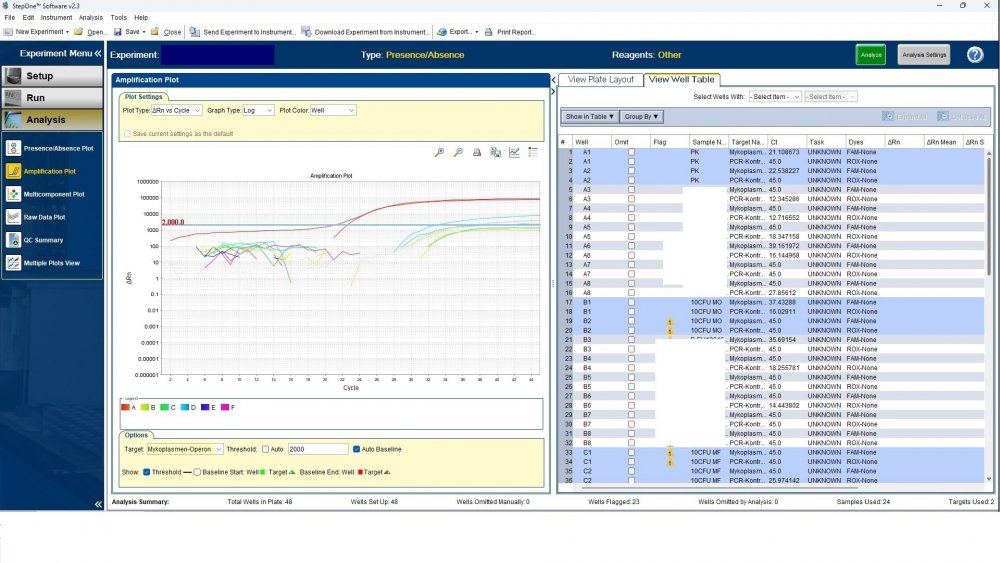
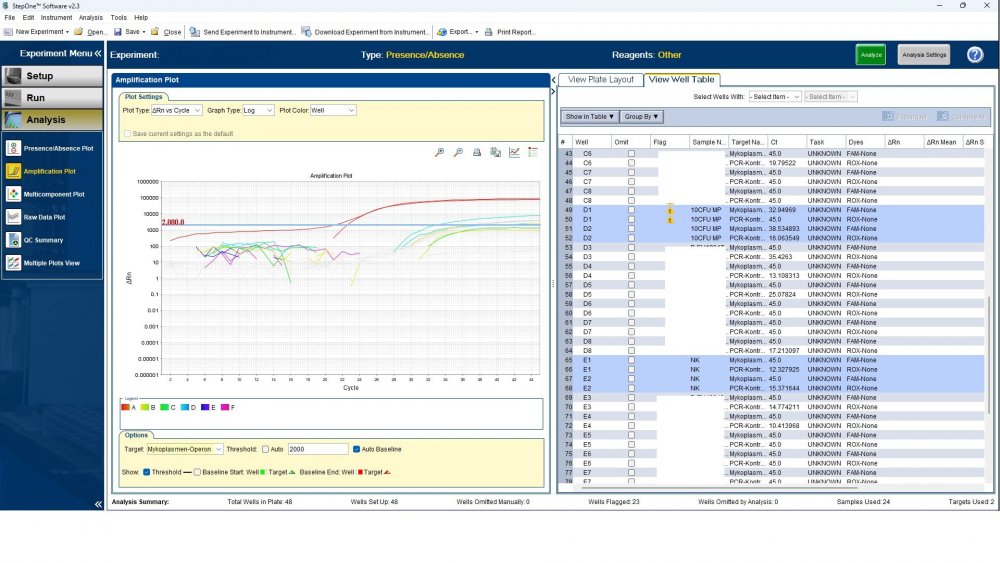
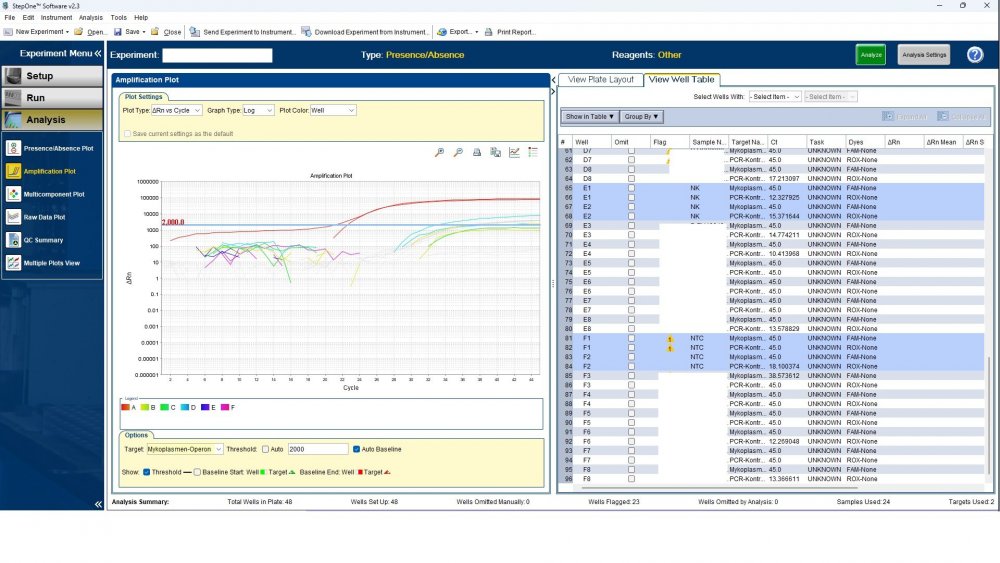
Maybe by explaining the steps of procedure, the issue will be clearer: - We buy cells infected with desired mycoplasma strains. - melting - counting (≥13x10^6) - Extraction - Measurement of CT genes extracted from prepared standards (usually at this stage, CTs show between 28 and 31) → allowed to be used - Approved for use as a standard (in the measurement of mycoplasma contamination of cell cultures) - Storage in the freezer -20 - My pipetting order is also clear: (two wells) A1, A2 = PK B1,B2 = Standard 1 (M. orale) C1,C2 = Standard 2 (M. pneumoniae) D1,D2 = Standard 3 (M. fermentans) E1,E2 = NK F1,F2 = NTC Correct results are obtained from wells A1, A2, E1,E2, F1,F2. Both samples of a standard are pipetted from a same microtube. (for example: microtube A containing 25 ml M. orale extracted gene, which is pipetted 10 ml in B1 and 10 ml in B2) - Two other standards in the same way. The strange thing is that when 2 samples are similar, how do they show 2 CTs with a big difference (sometimes one is less than 35 and the other is more than 35 and even 45). It is even more strange that I do not find any similarity in the results. (results are not interpretable). For example: One of the standards is outside the acceptable range. One of the wells is outside the acceptable range. One of the rows is outside the acceptable range. .... .... It seems that every time at least one of the 6 standards is out of scope and invalidates the work. As the result of the test may be valid by chance (10%). I have 2 devices and I see this happening in both. Thermo fisher recently recalibrated our PCR devices. Attention: We work under GMP principles, and all equipment and products are constantly calibrated and checked.
-
Cts value confuses me
What should I do to find out if the thresholds make sense or not? I use the Sartorius kit(FAM & ROX detection). The strange thing is that this kit used to work better with the approved method and for some time the number of unexplained errors became very high. I am looking for an idea to show why this happens. (Working with GMP principles is not easy to change, and the reason for the change must be provided)
-
Cts value confuses me
thanks CharonY, We work under GMP protocols. In this case, we have Mycoplasma infected cell lines (we buy). We count them. If there are more than at least 13 million cells, we extract DNA. After DNA extraction, we do PCR and according to the approved matrix, the standards should show a Ct less than 35 (According to the approved protocol). If we get approved Ct (≤35), we will be allowed (set free) to use them. What is interesting is that a total CT less than 30 is rarely obtained. But the main problem now: Why do we gain Cts above 35 and they appear randomly every time (90%)?!
-
Cts value confuses me
Hi everyone Mycoplasma detection has been a major challenge for me lately. I do cell culture and use qPCR to detect mycoplasma. In addition to controls(+,-) and NTC, I use 3 different strains of mycoplasma as standards (M. orale, M. pneumoniae and M. fermentans). Some Cts relating to standards are extremely high (sometimes 45!) for a few weeks. This is while I count the samples with a flow cytometer before DNA-Extraction and at least 13x10*6 cells are counted in it. The samples and wells in which Ct indicates greater than predicted (I consider ≤35 to be an acceptable Ct and I had before) appear to be entirely random. I have tested different modes, and the results are uninterpretable. We have two PCR devices, and the problem is the same in both. All samples, controls and standards are pipetted into two wells (double determination). Does anyone have any ideas?