raid517
Senior Members-
Posts
68 -
Joined
-
Last visited
Content Type
Profiles
Forums
Events
Everything posted by raid517
-
Well indeed, good point, there's an awful lot of them. Asexual reproduction/ conjugation etc. But aren't the chromosomes of the prokaryotes diploid? They may indeed not produce haploid gametes like the eukaryotes, but although they reproduce via binary fission, aren't their chromosomes still diploid?
-
OK, well you have my attention. Would you care to expand on your own comment on how 'the minority of organisms are diploid'? How would you count this to infer this outcome? I think you are correct and the authors wording is simply vague, and that what he means is that there are 4 copies of each of several important genes present in many of the jawed vertebrates. It is just a strange way to word it.
-
Hi, I am struggling with a phrase in a text book I am reading at a section discussing gene duplication (note this is not a question for amateurs). The phrase reads "the Genomes of many jawed vertebrates appear to have four diploid sets of many major genes." I have read this phrase several times, and for what ever reason it just isn't sinking in what it means? I mean I understand that the chromosomes of almost all living things are diploid in one way or another, but what does "four diploid sets of many major genes mean in reality? Would anyone care to expand on this?
-
Yeah that's a really great effort thanks. But I kind of had most of that already. The confusion is in what this guy in the video says at around 23:20 onwards, where he implies that one single DNA polymerase III is responsible for encoding both the leading and lagging strands. I just want to know if this is accurate, and if so how this can be the case? I suspect he's wrong, or at least I have been unable to source any video, or any animation showing how this might work. Instead most show two (or more) separate DNA polymerases encoding the leading and lagging strand simultaneously, all be it in a processive (for the leading strand) and (semi-processive) for the lagging strand) way. Can anyone explain if he is right how this is the case, or maybe find a video or animation of this process in action?
-
If you look at 23:25 onwards you will get a better idea of what he's saying. However every other video and animation I have seen say that there are two separate DNA polymerases that work to transcribe the leading and lagging strands. So which of these versions of events is accurate? Maybe its a dimer of some kind requiring the action of two joined (how though?) subunits of DNA polymerase? Surely this guy has got it wrong?
-
But what about DNA primase? During the replication of DNA small segments of primer RNA are laid down, so surely even though it's only for a short while, the nucleosome of Eukaryotes does contain a small amount of RNA?
-
Hi, in this video this guy appears to be implying that one single DNA polymerase molecule is responsible for the process of copying DNA. (I may however be reading him wrong.) He appears to imply that the DNA polymerase simply 'hops' back and forth across the two strands of the DNA molecule as DNA Helicase separates the two different strands. But surely it would make more sense and be more efficient if the replication process occurred using two separate polymerases? Since DNA polymerase 1 is progressive/semi-processive and DNA polymerase III is highly processive, wouldn't it be better to have these two enzymes do the two jobs separately at the same time? Also since there is 1 and 3, where does DNA polymerase 2 fit into the picture?
-
Hi could anyone please supply a step by step numerical breakdown of the processes involved in DNA replication? I mean I know some processes happen at the same time, but what's the general thrust of things? It would be really helpful if I could see something in a step 1, step 2, step 3 step 4 etc. type format. You see the problem is I've read about and watched loads of videos about each of the individual steps, but I'm having real trouble stitching it all together. Of particular confusion is in this video here: it shows the DNA molecule being copied into RNA first, presumably for later transcription back into DNA? However in this (really very good) online tutorial, it shows DNA being copied directly through a process of sem-conservative replication. So which is it? Or is it both? If so at which points do these processes occur? Also the initial video shows RNA-polymerase appearing to 'race along' the DNA molecule and unzip it to create an RNA template? But surely the RNA molecule doesn't move at all (or at least very little?) Isn't it the DNA molecule that gets fed through the RNA polymerase, a bit like film being fed through and old style cinema film camera?
-
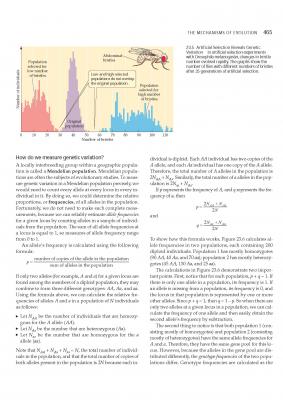
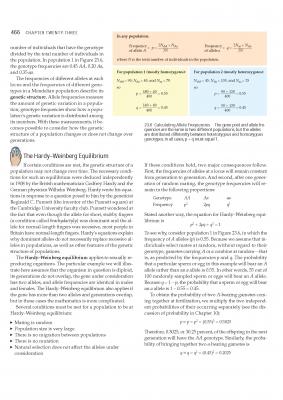
Hi in the following pages from a popular text book why in box 23.6 does it tell you that the population/sample size (in this case 200, or simply N), must be doubled in the equation to give 2N, to give p= 180+40/400, where 400 is equal to 2x N? In other words why do we need to double the population size to get the answer? I'm fine with the rest of the equation, so it's just this bit I'm stumped on. It might help if whoever replies to this reads both pages before responding to get an idea of what I'm driving at. This section is somewhat pre-Hardy Weinberg, but I will get around to that in due course once I have got the hang of this. As far as I understand it, the reason you divide the sum by 2N, is because if p= 2N(AA) + N(Aa) /2N then this means that both phenotypes are diploids? So to get the value of p (which is just the value a single allele within the population), you have to divide both sides of the equation by 2 to get the haploid value? This then allows you to ask how many copies of a single allele are present within any population? Can anyone confirm if this is correct?
-

What's the difference between ligands and cofactors?
raid517 replied to raid517's topic in Biochemistry and Molecular Biology
Thanks man. That's much clearer now. -

What's the difference between ligands and cofactors?
raid517 replied to raid517's topic in Biochemistry and Molecular Biology
OK, at a stab on the first question I suspect if ligands are non-covalently bonded molecules, then cofactors are covalently bonded? As for allosteric regulation the limit of what I understand about this can be found on the Wikipedia page about allosteric regulation. I don't really have time to write an essay based on what I read from this, as I'm supposed to be studying for an exam, and this isn't part of it. I just came across it when looking up something else. But the article doesn't make any clear distinction between the role of ligands and cofactors. Really I appreciate that you are trying to get me to think about this, but I just don't know what these distinctions are, so there isn't a lot of point in asking me to guess. Even if it were true that ligands are 'an umbrella term form non-covalently bound compounds to enzymes' where exactly does this leave us? Cofactors as far as I know bind to sites on the enzymes that are not the active site and cause the enzyme to have specificity in their activity in the presence of a given substrate. That is they can induce a conformational change in the enzyme to produce an induced fit mechanism that more tightly binds the enzyme to the substrate. A bit like a hand loosely grasping a ball at first, and then tightly grasping it, after it receives an appropriate signal to do so. But if you read the Wikipedia page on ligands, it implies (or seems to in places) that certain ligands can perform a similar function. To be honest it's all a bit of a muddle in my mind at the moment - and it would be useful if someone could help me separate the muddle out. Allosteric signaling is a mechanism whereby the presence (or lack thereof) of a given cofactor (or ligand?) within a medium can cause enzymes to speed up the rate of reaction (due to the above mechanism), or slow it down as appropriate. But it's entirely possible I am getting several related concepts confused. Some real world examples of where cofactors and ligands differ would be quite helpful I think. Also which of these is the most significant in terms of allosteric signaling and/or regulation? -

What's the difference between ligands and cofactors?
raid517 replied to raid517's topic in Biochemistry and Molecular Biology
Still not as clear as I would like. Do you have any examples to highlight the differences? Also do ligands have regulatory roles in the same way cofactors do? Are both, or only one active in allosteric regulation? -
OK I have just been thrown this word in one of the books I am reading, but with no real attempt made to explain it's meaning. But what in the context of chemotaxis is a "chemoeffector"? Does anyone know?
-
OK the title is a little misleading as I suspect that a Mega Dalton is simply a way of counting the overall size of something, when you don't know what all the constituent parts are? Is this correct? I mean looking it up on Wikipedia it says it says one Dalton is equivalent to the mass of one proton. Which is fair enough. So if a protein was one million Daltons in size (or one mega Dalton), that would just mean that it contains overall one million protons? (These are questions BTW). However the question is if this is correct, why is the Dalton used over any other measure in biology, how is it calculated and how is it denoted? Thanks!
-
I'm afraid it may take me the rest of my life to absorb what you just said. Nonetheless, I hope I get the opportunity to try. Thanks!
-
I still haven't got a clue what it's asking me to do. Wouldn't this be better in Biology, with biologists? It's not homework.
-
I'm completely stumped with this question. The problem is we were never told how to do this and the method for calculating this quantity doesn't seem to be in any of the supplied course literature. Does anyone know how to do this? Can you please show me?
-
"Strontium sulfate has a lower solubility in water at 25 °C than calcium sulfate. On the basis of this information, suggest what you would observe when dilute sulfuric acid is added to a solution of strontium nitrate with a concentration of 5 g per 100 g of water at 25 °C. Write down a balanced chemical equation that is consistent with your observation." Can anyone please help me work through this question? I'm stumped.
-
"De-excitation is not a gradual process" yet "It's not instantaneous." Lol, I think I probably just need to keep reading my textbooks. Thanks for trying anyway dude.
-
I guess we are going around in circles a little here, lol. If the de-excitation of electrons is truly instantaneous, then clearly (or seemingly), there would be no effectively measurable time period over which this would occur? It would either be in a high energy state, or a lower energy state and nothing in between. Actually this part kind of makes sense given that the principal quantum numbers of the electron shells can only have 'discrete' values. So one would not expect values in between n=4, n=3, n=2 and n=1 and so on. But the question still remains (for me) if the energies for the electron shells n=4, n=3, n=2 and n=1 etc are always specifically the same? That is (for example) can an electron lose (or convert) any of it's energy at all while it occupies the n=4 energy shell, before it drops to n=3, or n=2 etc? Or is the amount of energy the electron has while it occupies these shells always the same? If it cannot lose energy before dropping to subsequent shells, then what causes it to drop between one energy shell and the next, and for it to release a photon? I already accept that the attraction of the nucleus can pull the electron towards a lower state of potential energy, but surely if the photon was powerful enough to cause it to enter an excited state, either it must remain in that excited state, or lose some energy first before it can drop to a lower energy shell? I wonder if the answer to this question might simply be that it does lose energy - in the form of a photon, lol. Also it was you I believe who said that photons are both created and destroyed, but then when I ask how they are created and destroyed, you say that you know of no mechanism for this and that it cannot be tested. Very confusing... I do know that photons can last for a very long time, in some cases (in terms of the cosmic background radiation), for as long as the Universe has existed. So they may not always be destroyed. But surely if some mechanism exists for their creation and destruction, we should at least be capable of knowing what it is? Just saying that a photon is released when an electron drops from one energy state to another and is absorbed when a photon of just the right energy excites an electron to enter a higher energy orbital, does not explain how a photon is absorbed, or emitted, it just says that it is. I agree. I don't expect I'm going to get any vastly useful answers here. Small simple hints perhaps. It's just that as I read my textbooks a billion questions like this spring to mind, but perhaps I will have to simply shelve many of them until I have finished some of my reading.
-
This is all very vague. What does 'discretely' mean in this context? Also what does 'you can't tell one photon from another' mean? Do you mean that the energy of the photon transmitted will be the same as the energy of the photon that is absorbed, so you can't tell the difference between them? The last part too is a little unclear, how are photons created and how are they destroyed? I think it's very difficult to lean on an internet forum, lol.
-
Yes that's rather what I said previously. But the rest of the questions are still interesting to me if anyone has an answer?
-
I'm still trying to visualise the full actual sequence in my head. The photon strikes the electron, the electron becomes excited and is raised to a higher energy level. A 'period of time' elapses (given that this will be extremely short), which is sufficient for the electron to lose energy. The electron (now having lost energy), drops to a lower energy state, as the force supplied by the photon to raise it to it's excited state is gradually overcome by the force of attraction of the nucleus, and finally the electron releases a photon. A number of questions emerge from this. Firstly is this an accurate model of the process? Secondly if so is the photon that's emitted always exactly the same energy and wavelength of the photon that was absorbed? Thirdly, if so is the photon that was emitted the 'same' photon that was absorbed? Or for example is the photon that is emitted simply a result of energy transfer, where the kinetic energy of the excited photon is gradually lost and transferred to light energy, thereby causing a photon to be emitted? If so where did this 'new' photon come from? If it is the same photon that was absorbed, then why is the electron able to hold on to it in it's excited state and why does it release it in it's non-excited state? Is there some kind of force of attraction between a photon and an electron in it's excited state? Also in relation to the third point, are photons ever destroyed, or are they simply absorbed and re-emitted later? Sorry for all the questions, it just seems like an interesting topic and it would probably be useful if I could get a firmer grasp of it.
-
I'm not sure if this is still really explaining it. I think we are (roughly!) on the same page that an electron will tend to emit a photon and not contentiously store energy from succeeding photons, because there is 'enough time' between photons striking it for it to lose energy (although maybe this is too simplistic?), but the part where it gets a little fuzzy for me is why it emits this energy in the form of a photon and not in some other way? However I also suspect that the answer to this might be so complex that it's beyond me right now lol. Just saying because something can happen it will happen is interesting, but maybe a little too philosophical for me at this point. So what is this force in terms of the atom (say hydrogen) and it's electron? If it was a ball the force pushing (or in this case pulling) the ball towards it's state of lowest potential energy would of course be gravity.